Simulation with yeast_model.fig
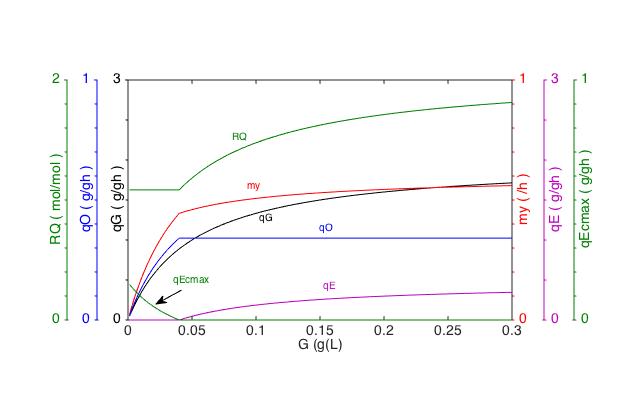 G:
Glucose concentration; qE: Spec. ethanol production rate; qO:
Spec. oxygen consumption rate; my: Specific growth rate
qG: Spec. glucose consumption rate; RQ: Respiratory quotient; qEcmax: Max ethanol uptake rate |
|
Kinetic
model of overflow metabolism in Saccharomyces cerevisiae.
In a process where both ethanol and glucose are present at glucose concentrations below the critical value, ethanol is re-assimilated at rate (qEcmax*E/(E+Ke)) that is proportional to the “free capacity”, i.e. the difference between qOmax and the actual qO. Therefore qO is equal to qOmax as long as ethanol is present in the process. For further illustration of the model check the simulation with yeast_fb.fig, which uses this model for simulation of a fed-batch process where ethanol is first produced but later on in the process consumed. A
simulation of this type requires stoichiometric analysis. |
For information about model equations and parameters: Open the SIMSPEC-file yeast_model.fig, available in the SimuPlot toolbox.