Simulation with yeast_fedbatch.fig
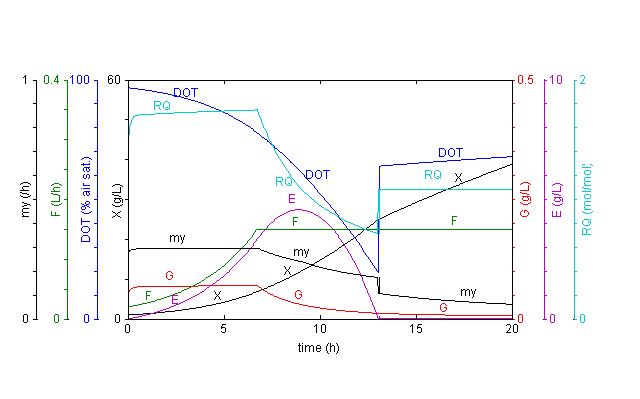 X:
Biomass concentration; G: Glucose concentration; DOT: Dissolved
oxygen tension; E: Ethanol concentration
my: Specific growth rate; F: Feed rate; RQ: Respiratory quotient. |
|
Dynamic
simulation of a fed-batch process with Saccharomyces
cerevisiae. Also under fully aerobic conditions does this
organism produce ethanol due to overflow metabolism, if the
glucose concentration is too high. To avoid this, the process
is usually started with a low (limiting) concentration of
glucose which is kept approx. constant by an exponential feed.
The specific growth rate (my) then becomes approx. equal to the
exponent of the feed profile. Due to the exponential growth DOT
declines and before it becomes too low the feed is switch to a
constant flow. For further illustration of the model see the SimuPlot simulation with yeast_model, which plots the specific rates against glucose concentration in Monod type plots. A
simulation of this type requires stoichiometric analysis. |
For information about model equations and parameters: Open the SIMSPEC-file yeast_fedbatch.fig, available in the SimuPlot toolbox.