Simulation with Sgradient_bead.fig
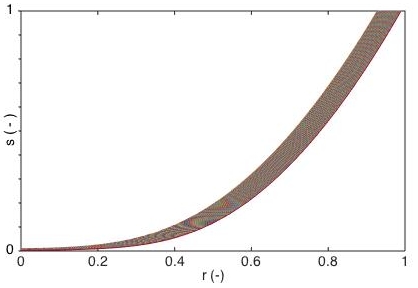 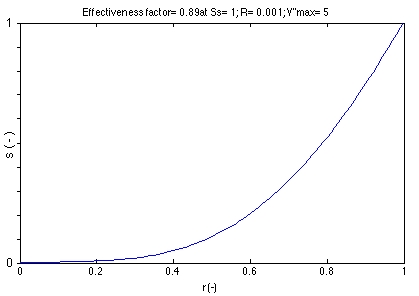 The
dimensionless substrate concentration was calculated as function of
dimensionless distance (r) from the center of a homogeneous biocatalyst. |
|
The
substrate concentration gradient in a homogeneous bead with cells or
enzyme consuming the substrate can be described with a second order
differential equation that, after conversion to dimensionless
form, can be solved numerically.
The
theory
of substrate diffusion limitation and its effect of the effectiveness
factor (diffusion limited/ non-limited reaction rate) is included in
Fermentation Process
Engineering, 2nd ed. (will become available for downloading from
www.enfors.eu). |
For information about model equations and parameters: Open the SIMSPEC-file Sgradient_bead.fig, available in the SimuPlot toolbox.